Rhesus Macaque (Macaca mulatta) Contig Mappings onto Human via Pash
(Version: Rhesus Mmul_0.1 Assembly - Human NCBI Build 34)
PGI Method Explanation
Rhesus PGI Paper
Search for Rhesus BACs covering a Human gene,
or other area of interest:
- Enter the RefSeq Gene Symbol or gene name.
- You will see the Rhesus BACs covering your gene, if any, as a Genboree visualization.
- You can also try searching using other "Keywords" (eg, accession numbers, cytobands, etc)
- You can also search for Rhesus BACs directly by entering their CHORI names here.
Genboree-Browsable Data
The VGP image below indicates the distribution of our 16,495 Rhesus CHORI-250 BACs mapped to the Human genome NCBI Build 34 via the PGI method (blue track). Putative rearrangements between Rhesus and Human, inferred from the BAC mapping results, are indicated in the red track. The numbers above the tracks indicate the number of Rhesus BACs mapped to each Human chromosome. The visualization is clickable and provides links to view the PGI data within Genboree.
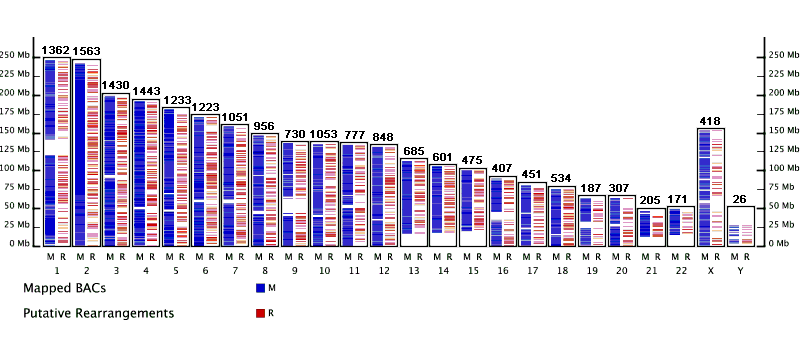
To use the VGP Image:
- Click on any chromosome to view the indices in Genboree.
- Once in Genboree, you can refine your region of interest using the various navigation features.
Raw Data Download
We also provide the detailed raw mapping data for each of the Rhesus BAC mappings.
- Download the raw data for all BAC mappings as a tab delimited file.
NOTE: Contact CHORI if you wish to obtain a BAC for in-depth sequencing.
Pool Read Download
Pool Project ID:
- Enter a 4-letter pool project id to access the pool reads in NCBI Trace Archive.
- Pools contain reads from multiple Rhesus BACs.
- You may want to examine the array layout files below for lists of pool ids and their BAC composition.
Experiment-Related Files
- For each Rhesus array-pair, we used the same transversal design relation between the original array layout and the shuffled array layout:
- 1st 48x48 BAC Array-pair:
- 2nd 48x48 BAC Array-pair:
- 3rd 48x48 BAC Array-pair:
- 4th 48x48 BAC Array-pair:
- 5th 48x48 BAC Array-pair:
- 6th 48x48 BAC Array-pair:
- 1st 96x96 BAC Array-pair: